Article Text
Abstract
Background: The genetic basis of variation in human cognitive abilities is poorly understood. RIMS1 encodes a synapse active-zone protein with important roles in the maintenance of normal synaptic function: mice lacking this protein have greatly reduced learning ability and memory function.
Objective: An established paradigm examining the structural and functional effects of mutations in genes expressed in the eye and the brain was used to study a kindred with an inherited retinal dystrophy due to RIMS1 mutation.
Materials and methods: Neuropsychological tests and high-resolution MRI brain scanning were undertaken in the kindred. In a population cohort, neuropsychological scores were associated with common variation in RIMS1. Additionally, RIMS1 was sequenced in top-scoring individuals. Evolution of RIMS1 was assessed, and its expression in developing human brain was studied.
Results: Affected individuals showed significantly enhanced cognitive abilities across a range of domains. Analysis suggests that factors other than RIMS1 mutation were unlikely to explain enhanced cognition. No association with common variation and verbal IQ was found in the population cohort, and no other mutations in RIMS1 were detected in the highest scoring individuals from this cohort. RIMS1 protein is expressed in developing human brain, but RIMS1 does not seem to have been subjected to accelerated evolution in man.
Conclusions: A possible role for RIMS1 in the enhancement of cognitive function at least in this kindred is suggested. Although further work is clearly required to explore these findings before a role for RIMS1 in human cognition can be formally accepted, the findings suggest that genetic mutation may enhance human cognition in some cases.
- LBC, Lothian birth cohort
- LOD, logarithm of odd
- SNP, single-nucleotide polymorphism
- VIQ, verbal IQ
Statistics from Altmetric.com
A genetic contribution to variation in human intelligence is well established, but the identities of the genes responsible remain elusive. Many mutations are associated with impaired cognition:1 no definite genetic causes of enhanced cognition are established,2 and there are no known cognition-enhancing “gain-of-function” mutations in genes otherwise associated with cognitive impairment. Therapeutic possibilities deriving from the discovery of any such genes or variants are potentially important: cognitive decline reduces the quality of life,3 and low intelligence test scores are associated with increased morbidity and shorter life-span.4 Accelerated evolution of genes subserving neurodevelopment figures in molecular explanations of the advance of the human nervous system: many of the identified genes regulate brain size and behaviour, some encoding critical synaptic proteins.5
To identify genes influencing human brain development and function, including cognitive function, we use a paradigm evaluating cerebral structure and function in individuals with known mutations in genes co-expressed in the lineage-sharing eye and brain, ascertained by their obvious ocular phenotype, but in whom a neurological phenotype was not fully appreciated. Using this paradigm, we demonstrated roles for the genes PAX6, PITX2, SOX2 and OTX2 in human brain development, cognitive function and memory.6,7,8,9,10,11
We now report on the functional and structural effects of mutation in the eye- and brain-expressed gene RIMS1, through the study of individuals from a family already reported to have retinal dystrophy caused by RIMS1 mutation.12,13 To our knowledge, this is the only family so far reported with such a mutation: the eye phenotype is homogeneous in the family, and has been documented in detail.13 The orthologous murine Rim1α encodes a synaptic active-zone protein necessary for preserving the normal probability of synaptic neurotransmitter release and for long-term presynaptic potentiation.14,15Rim1α is also expressed in retinal ribbon synapses.16 Mice lacking Rim1α protein show severely impaired learning and memory.17 In our kindred, RIMS1 mutation (Arg844His) causes a late-onset dominantly inherited cone–rod dystrophy (CORD7; OMIM 603649), leading to varying degrees of visual loss starting from the third decade onwards.13 Because RIMS1/Rim1α is also expressed in the brain, we hypothesised that this RIMS1 mutation would produce a structural and functional neurological phenotype.
MATERIALS AND METHODS
Ethics
The study of the kindred was approved by the Research Ethics Committees of the Institute of Neurology, National Hospital for Neurology and Neurosurgery and Moorfields Eye Hospital, and The Lothian Birth Cohort study was approved by the Lothian Research Ethics Committee. All participating individuals provided written informed consent. Human embryonic/fetal material was studied by the Human Developmental Biology Resource at the Institute of Child Health with full ethics approval.
Cognitive tests applied to kindred
The neuropsychological tests used had been used in an earlier investigation of individuals heterozygous for PAX6 gene mutations.7 All the standardised tests selected always require verbal interaction only, accounting for variable vision impairments: subjects with preserved vision (both mutation carriers and unaffected kin) were thus not disadvantaged. Details of the tests used are given in the supplementary material (available online at http://jmg.bmj.com/supplemental). Intellectual level, executive and memory functions were tested.
Brain imaging
Details of the high-resolution MRI are provided in the supplementary material (available online at http://jmg.bmj.com/supplemental).
Population genetics
Subjects
All subjects were selected from the Lothian birth cohort of 1921 (LBC1921) who are survivors from the Scottish Mental Survey of 1932.18 On 1 June 1932, a valid mental ability test, a version of the Moray House Test No 12, was administered to almost all Scottish children who were born in 1921 and attending school that day (n = 87 498). The mean (SD) age at re-test was 79.1 (0.6) years. The following inclusion criteria were adopted for each subject: (1) Moray House Test score at age 11 years had to be available; (2) there had to be no history of dementia; (3) Mini-Mental State Examination score was ⩾24; and (4) genotyping had to be successful for RIMS1. This gave a total of 469 subjects (278 women).
Cognitive testing
Details are provided in the supplementary material (available online at http://jmg.bmj.com/supplemental): Mini-Mental State Examination, Moray House Test, Raven’s Standard Progressive Matrices, verbal fluency, logical memory and the National Adult Reading Test were measured. The g factor, as a measure of general intelligence, was created by principal component analyses of some of these measures: a single component accounted for 53.5% of the total variance.
Tag selection
Genotypes of RIMS1 single-nucleotide polymorphisms (SNPs) with a minor allele frequency of at least 0.05 were downloaded for CEPH trios from the HapMap database, across a 750 kb region of DNA comprising the RIMS1 gene, with 100 kb upstream and downstream (242 SNPs). The region was divided into seven sections of high linkage disequilibrium using the pairwise D′ plot. Haplotypes were inferred for each section (TagIT: http://www.genome.duke.edu/resources/computation/software). These haplotypes were then used, within TagIT, to select tagging SNPs for each section, ensuring that the average-locus haplotype r2 value of each section within the gene was ⩾0.8. Genotyping and statistical analysis were carried out as detailed in the supplementary methods (available online at http://jmg.bmj.com/supplemental).
RIMS1 sequencing in LBC1921
PCR mix composition, primer sequences, cycling protocols and sequencing details are available on request.
RIMS1 expression studies
RIMS1 mRNA expression was characterised in random-bred CD1 adult mouse brain and eye. The distribution of RIMS1 mRNA was studied in the human brain and eye at the Carnegie stage 21 and the fetal stage F1. Riboprobe generation and in situ hybridisation are detailed in the supplementary material (available online at http://jmg.bmj.com/supplemental).
RIMS1 evolution
Sequencing
Human, chimpanzee, rat and mouse sequences were obtained from GenBank (acc nos NM_014989, XM_527433, NM_052829 and NM_053270, respectively). Sequences were aligned using BioEdit, and DNA mutations were identified. RIMS1 sequences for macaque and squirrel monkey were sequenced from genomic DNA. PCR primers were designed using the human RIMS1 genomic sequence as a template, and the presence of PCR product was confirmed by gel electrophoresis. Sequencing was conducted using the BigDye terminator kit and sequences read on an ABI 3700.
Calculation of KA/KS
KA and KS were determined according to the following definitions:
KA—number of non-synonymous substitutions per non-synonymous site
KS—number of synonymous substitutions per synonymous site
Sequences were aligned using BioEdit, and only portions for which sequences were available for both species were included in the calculations. The amino acid content was ascertained for both species and the average number of synonymous and non-synonymous sites was calculated according to the following:
Non-synonymous sites = non-degenerate sites + 2/3 two-fold degenerates.
Synonymous sites = four-fold degenerates + 1/3 two-fold degenerates.
The average number of synonymous and non-synonymous sites was used for the calculation of KA and KS. The significance of KA/KS between primates and rodents was assessed using a simple χ2 evaluation.
RESULTS
Cognitive function is enhanced in individuals with RIMS1 mutation
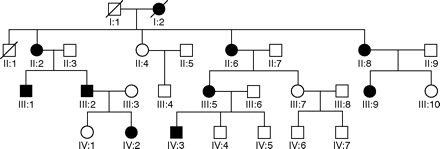
The pedigree is illustrated in fig 1. Eight (of nine total) affected and seven (of nine total) unaffected adults were assessed. The verbal IQ (VIQ) is a composite measure generated from a range of tests. VIQ was pro-rated from the Wechsler Adult Intelligence Scale (revised) Vocabulary, Digit Span and Similarities subtests.19 VIQs in affected individuals (table 1) were all above average, with quotients ranging from the 75th (high average ability) to the 99th centile (superior ability). Centile levels for subjects II:2 and II:6 are probably underestimates, as normative data are available up to 75 years only. Some pro-rated VIQ scores, for example 150 in subject III:1, reflect a ceiling effect. On measures of memory and executive skills, performance levels were more variable, with some scores falling below average. Normative data available for these measures were based on smaller sample sizes than for VIQ and, with the exception of semantic fluency, were not available for older age bands.
Scores and centile performance (age corrected) for verbal IQ, memory and executive skills tests
The kindred studied. Filled symbols represent affected individuals, unfilled symbols represent unaffected individuals. Note that the kindred is an extension of that presented by Johnson et al12 and Kelsell et al,20 reordered by age of generation II (and thus does not seem to be identical to previous presentations).
The identification of RIMS1 (and its mutation) as a causal factor in this kindred might usually be undertaken using linkage analysis, both for the previously reported retinal phenotype (maximum logarithm of odd (LOD) score 3.61 at θ = 0) and for the phenotype being considered here, VIQ. For VIQ, fewer informative members were tested: using a high IQ phenotype classified as >1 SD (15 verbal IQ points) above the mean) to calculate linkage gives a maximum LOD value of 1.92 at a recombination fraction (θ) of 0.2, which is highly indicative of linkage. This analysis, however, imposes a somewhat arbitrary interpretation of the VIQ phenotype, and does not allow to be taken into account other cognitive information available.
Fortunately, the process of Mendelian inheritance allows a further robust test of the causality of the Arg844His mutation to be performed, avoiding the usual problems of confounding from other variables that might aggregate in a familial fashion but might be unlinked to the mutation. To test the relevance of the mutation, affected and unaffected first-degree sibs in each generation (II:2, II:6, II:8 vs II:4; III:5 vs III:7; IV:3 vs IV:4, IV:5) underwent identical cognitive testing (table 1). The mean age of affected members (53.8 years) was higher than that of unaffected members (41.1 years), acting conservatively against any superior cognitive performance in the affected group, given the known effect of age.21 We tested the significance of the association between affected status and every measured cognitive phenotype following a randomisation approach that accounts for the kindred structure.22RIMS1 genotypes are randomised across the pedigree, following Mendelian inheritance, while keeping the phenotype values of each individual fixed. The null hypothesis therefore allows for the possibility of familial effects on phenotypes that are unlinked to RIMS1, and any significant departure from the null hypothesis can only be ascribed to an effect linked to RIMS1. The 11 measured phenotypes were vocabulary (subcomponent of VIQ), digit span (subcomponent of VIQ), similarities (subcomponent of VIQ), phonemic fluency, cognitive estimates, immediate story recall, delayed story recall, immediate verbal learning, delayed verbal learning, semantic fluency and Hayling test.
The perfect observed association between the affected status and RIMS1 mutation allowed us to infer a dominant Mendelian disease model, and also to infer the genotypes of all founders and marry-ins. We simulated genotypes, and then the affected status of all individuals in the pedigree based on Mendelian inheritance from the known founders, and for each normalised phenotype we calculated the difference in means between the affected and non-affected indivduals (keeping phenotype values fixed among individuals). To address multiple testing issues, we generated a single score for the overall difference between the affected and non-affected groups. We first normalised all phenotypes to a common variance scale, dividing by the standard deviations estimated from a disease-control group of 15 visually impaired individuals with mutation in PAX6,7 so all phenotypes had equal weight. We then used the Euclidean distance of the 11 difference-in-means values. To control for possible ascertainment bias, we further conditioned on the observed number of the affected and non-affected individuals in the pedigree by rejecting any pedigrees from the Monte Carlo process that did not match this. We carried out 10 000 simulations under these conditions.
The single score for the overall difference in all mean phenotype values between the affected and non-affected subjects was significant (p = 0.006). For individual phenotypes, all except those for immediate story recall, delayed story recall and semantic fluency had p values <5%: vocabulary (p = 0.014), digit span (p = 0.020), similarities (p = 0.039), phonemic fluency (p = 0.050), cognitive estimates (p = 0.012), immediate story recall (p = 0.112), delayed story recall (p = 0.087), immediate verbal learning (p = 0.048), delayed verbal learning (p = 0.042), semantic fluency (p = 0.220) and Hayling test (p = 0.010). As expected, given that the three subphenotypes of VIQ were all significant, when we separately tested VIQ itself this was also significant (p = 0.014).
We determined to what extent the coincidence of these associations is due to the natural correlations among these phenotypes. Table 2 presents the correlation coefficients of the 11 phenotypes and VIQ, in the disease-control sample of 15 individuals with PAX6 mutation.7 Given that the three subphenotypes of VIQ were all significant, when we separately tested VIQ itself, this was also found to be significant (p = 0.014): although this particular result reflects a part–whole correlation, it is of note that all the 11 cognitive phenotypes did not naturally correlate with each other, as assessed in the disease-control group.7 Phonemic fluency, verbal learning and semantic fluency were highly correlated with VIQ, and it is unclear whether the significant results observed in some of these variables were simply due to correlation with VIQ. However, the phenotypes of cognitive estimates and Hayling test were poorly correlated with VIQ, the other variables and each other, possibly suggesting that multiple facets of mental processes were being affected independently by the mutation.
Correlation coefficients of phenotypes in disease-control (PAX6) sample
There was no history or genetic evidence for inbreeding (number of markers scored = 16, heterozygous average for 14 members of kindred = 11.6 (72%); equivalent scores for five “married-ins” = 11.4 (71%); data from ref 22, omitting IV:4–7).
To address the specific, but unlikely,23 possibility that visual impairment might enhance function in other domains, we compared identical measures from the disease-control cohort of 15 individuals (mean age 36 years, range 17–54 years; eight women), all of whom had congenitally symptomatic PAX6 mutations.7 The PAX6 subjects had earlier onset of visual symptoms and more severe central visual impairment at the time of cognitive testing than the RIMS1 subjects (supplementary table 1 available online at http://jmg.bmj.com/supplemental). The mean VIQ of the PAX6 subjects was 103. The PAX6 subjects as a group do not differ from the RIMS1-unaffected individuals: cognitive measures for both groups fall within the normal ranges (table 3). Despite their more severe and earlier-onset visual impairment, the PAX6-affected individuals performed significantly less well than the RIMS1-affected individuals (VIQ, Mann–Whitney U test p <0.0005). In addition, in three of eight affected RIMS1 individuals, visual acuity was normal (6/6) or near normal (6/9) at the time of cognitive assessment.
RIMS1 and PAX6 group cognitive test scores
Brain structure in individuals with RIMS1 mutation
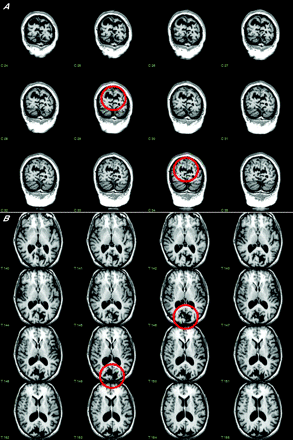
In seven affected members of the kindred, high-resolution brain MRI was obtained. In two, a mother (II:2) and son (III:1), brain abnormalities were apparent on inspection (fig 2A, B). Cortical grey matter was preserved around widened cerebrospinal fluid spaces: these changes would be compatible with a bilateral parasagittal polymicrogyric malformation.
Serial reformatted T1-weighted magnetic resonance images from subject II:2, shown in (A) coronal and (B) axial planes. Note the widened cerebrospinal fluid spaces around an area of cortical malformation, shown encircled in selected images. Detailed inspection in three dimensions suggested that the malformation was most probably an area of polymicrogyria.
RIMS1 genetic variation in other cone–rod dystrophy patients and a population cohort
We sequenced the mutation-containing RIMS1 exon 13 in a panel of 50 unrelated individuals with autosomal dominant cone–rod dystrophy, but did not detect any mutations. Common variation in RIMS1 (uncorrelated with the rare Arg844His mutation) did not influence cognitive function in LBC1921, for either genotype or haplotype (see supplementary tables 2 and 3 available online at http://jmg.bmj.com/supplemental). To determine if mutation in RIMS1 might account for the upper extreme of performance on cognitive measures, the entire RIMS1 gene was sequenced in the top-scoring 5% of the LBC (24 individuals). Only one, previously unreported, SNP was found, in residue 592, exon 9: it was synonymous, conserving a glutamic acid residue in an unremarkable region of the gene.
RIMS1 brain expression in human fetal and adult brain
In situ hybridisation with an antisense RIMS1 probe in fetal stage 1 human embryos revealed RIMS1 expression in the outer plexiform layer of the developing retina, but not in the optic nerve, as expected (figs 3A–C, F). Cerebral expression was seen in the ventricular zone, developing cortical plate, thalami and hippocampal anlage (fig 3C–E, G). Telencephalic RIMS1 hybridisation signal was detectable from 57 days after-conception onwards, but not before. In adult human hippocampi from patients who had undergone therapeutic resective surgery for refractory temporal lobe epilepsy due to hippocampal sclerosis, RIMS1 expression was observed at least in the granule cell layer and CA1 (fig 3H): in the absence of other suitable adult brain material, expression elsewhere cannot be excluded.
In situ hybridisation for RIMS1 mRNA in human embryonic eye (A) from Carnegie stage 21. The purple labelling represents expression of RIMS1, found as expected in the developing retinal pigment epithelium, shown at higher power in (B). (A) and (B) are sagittal images. In (C), a tilted coronal section through the developing human head at fetal stage 1 shows expression in the developing cortical anlage (D, higher power), hippocampal anlagen (E, arrows) and retina, but not in the optic nerve (on) (F). Expression is present in both the subventricular zone and the cortical plate (G, arrowheads). In adult human hippocampus (H), from a patient with temporal lobe epilepsy, expression (purple labelling) is seen in small cells in the (neuron-depleted) dentate granule cell (GCL−) layer and in the CA1 sector (CA1−), compared with sections labelled with the sense probe as control (GCL+, CA1+).
RIMS1 evolution
The value of the ratio of non-synonymous (KA) to synonymous (KS) nucleotide substitution is a measure of selection pressure on a gene: overwhelmingly, non-synonymous alterations are deleterious, and subject to negative selection. Rarely, non-synonymous changes improve protein function and are subject to positive selection, with an elevated KA/KS. To determine whether RIMS1 has been subject to positive selection, we estimated RIMS1 KA/KS for humans compared with chimpanzee and macaque, considered against rate change in a rodent divergence (rat–mouse) following Dorus et al.5 We were unable to demonstrate accelerated evolution of RIMS1 in the primate lineage (see supplementary material available online at http://jmg.bmj.com/supplemental).
DISCUSSION
Few individual genetic factors underlying the heritability of human intelligence are known.2 Mutations in many genes, inherited in Mendelian fashion, are associated with reduced intelligence,1 often associated with other problems. Notably, recent variation in two such genes (MCPH1 and ASPM) has been shown to be subject to strong positive selection, indicating ongoing adaptive evolution of the human brain24,25 and emphasising the importance of population genetic analyses of genes mutations in which affect human cognition. Quantitative trait analyses in populations with learning disabilities also suggest that genes with Mendelian effects, when mutated, are broadly candidates to influence normal variation in learning abilities in the general population.26 To our knowledge, no human genetic mutations have yet been identified that enhance cognitive function.
We show that a mutation in RIMS1 is associated, in the only reported kindred with any RIMS1 mutation, with significantly enhanced cognitive function in at least the verbal (likely to be related to general ability, g) and executive domains. RIMS1 is an excellent candidate gene to influence cognitive function. Rim1α protein regulates synaptic-vesicle fusion, and interacts with several other active-zone molecules, including α-liprins, Munc 13-1, CAST, SNAP-25, synaptotagmin and 14-3-3, generating a protein scaffold in the presynaptic terminal.27 Rim1α greatly enhances neurotransmitter exocytosis in a Rab3-dependent manner.28 Absence of Rim1α protein leads to severe impairment of learning and memory.17
Could the RIMS1 mutation be a chance finding, unrelated to the eye or cognitive phenotype? The intrafamilial distribution of cognitive measures argues that the detected mutation is most probably causative, especially as it segregates with both the eye phenotype (which becomes clinically symptomatic) and the cognitive enhancement. Co-mingling in each outbred generation of mutation-carrying and wild-type sibs each with respective enhanced or normal cognitive phenotype and the respective co-segregating impaired or normal visual phenotype renders extremely unlikely the possibility of an intrafamilial founder effect unrelated to the RIMS1 mutation, as supported quantitatively by our modelling, beyond linkage analysis alone.
We have not demonstrated a molecular basis by which the R844H substitution in RIMS1 could enhance cognition. The possibility remains that there is a mutation (or mutations) in another gene (or genes) in the region between markers D6S430 and D6S162512, which is (or are) responsible for both the retinal and cognitive phenotypes, or the retinal and cognitive phenotypes separately. Within this region, there are at least 17 other genes. Two of these, IMPG1 and MYO6, were considered good candidates for the eye phenotype alongside RIMS1 on the basis of function and pattern of expression, and were screened by Johnson et al.12BAI3 was subsequently screened (Cottrill and Hunt, unpublished data): no mutations were found in any of these other three genes. Yet other genes, such as KCNQ5, where there is no evidence for retinal expression, have not been examined, and would not be candidates for the eye phenotype, but could be for the brain phenotype. However, we consider it more likely on the basis of the available evidence, including biological plausibility,14,15,17 that the R844H mutation in RIMS1 explains both the eye phenotype and the cognitive phenotype, with a lower LOD score for the cognitive phenotype as this is more genetically complex and the mutation probably less penetrant.
One interpretation of our findings is that the RIMS1 mutation in this kindred alone (ie, with this genetic background and environmental setting) is permissive of a gain of verbal cognitive abilities in response to impaired visual function, rather than being directly causative. This is inherently unlikely, especially in this kindred, as with increasing age higher cognitive abilities are usually associated with better, rather than defective, sensory processing.23 In the RIMS1 kindred, development of visual symptoms occurs later than the age at which values of cognitive measures (eg, NART, VIQ) are set—for example, subject III:2 did not develop symptomatic retinal dystrophy until the age of 42 years, but had VIQ 146 at the age of 49 years. In addition, three mutation carriers in the kindred had normal visual function at the time of cognitive assessment. Additionally, visual impairment due to PAX6 mutation in unrelated subjects with earlier and more severe loss of central vision was not associated with any gain in VIQ. Therefore, the data suggest that the observed genetic change is a gain-of-function mutation leading to increased performance in specific cognitive domains in affected humans, irrespective of other genetic background and unrelated to visual impairment. The RIMS1 mutation studied may affect other cognitive domains—the paradigm we applied is biased to tests requiring only auditory presentation and verbal response, and verbal, not visual, processing, and was used for subjects we suspected of having cognitive impairment, not cognitive enhancement. Thus, only a limited cognitive assessment was undertaken, in line with our previous reports. We note that, undoubtedly, other genes must also contribute to cognitive scores in this kindred.
The RIMS1 gene shows extensive organ-specific alternative mRNA splicing.12 Thus, although the mutation lies in the protein C2A domain, and is shared by the eye and brain isoforms, nevertheless the ocular neurophysiology in the kindred13 need not reflect mutant brain RIMS1 protein function. Lack of homogeneity within a kindred for the functional consequences of gene mutation has previously been noted.6 Our qualitative structural findings also implicate RIMS1 as a candidate gene for bilateral parasagittal polymicrogyria. Brain malformation is not a prerequisite for the cognitive phenotype in this kindred, and, indeed, when cognition is altered in patients with polymicrogyria, it is usually impaired.
RIMS1 is a plausible candidate because of data from animal studies discussed earlier, and also in terms of the pattern of expression in humans. Thus, human RIMS1 mRNA is detectable from an early age not only in retinal, but also in cerebral, anlagen (fig 3) and adult human hippocampus (fig 3H). RIMS1 protein is more abundant in phylogenetically newer brain regions than in older regions.28 However, we were not able to demonstrate accelerated evolution of RIMS1 in the primate lineage, nor were genes encoding RIMS1-interacting molecules primate-fast outliers in a previous analysis.5
The population genetic data suggest that RIMS1 may not have a major role in normal variation of the studied cognitive measures in a Western European population. Any evolutionary advantage of this particular mutation could be counterbalanced by the concomitant severe visual phenotype, albeit late onset. Although population genetic studies can provide useful, or even conclusive, evidence that a gene and its encoded protein have a biological function, negative population genetic studies do not exclude such a role. The genetics of Parkinson’s disease illustrate this well: rare, early-onset Mendelian cases may be caused by mutation in PINK1,29 but common variation in PINK1 does not influence the risk of common, sporadic Parkinson’s disease.30 Further work is clearly required before a role for RIMS1 in human cognition can be formally accepted. However, even if the observed RIMS1 mutation turns out to be a private mutation enhancing cognition in affected members of this kindred only, the observation remains important because as the first such mutation identified in humans, it raises the possibility that genetic mutations may lead to increased, as well as reduced, cognitive function in man.
Acknowledgments
We thank the kindred. The work was supported by the Guide Dogs for the Blind Association (ATM, DMH), British Retinitis Pigmentosa Society (DMH, ATM), the UK’s Medical Research Council UK (SLF) and the National Society for Epilepsy, UK (SMS, PJT). The Lothian Birth Cohort 1921 was supported by the UK’s Biotechnology and Biological Sciences Research Council. IJD is the recipient of a Royal Society-Wolfson Research Merit Award. Human embryonic and fetal material was provided by the Medical Research Council/Wellcome Trust-funded Human Developmental Biology Resource.
REFERENCES
Supplementary materials
Files in this Data Supplement:
Footnotes
-
Published Online First 19 January 2007
-
Competing interests: None declared.